Early embryonal development of Drosophila
Precision and scaling in the pattern formation in the early embryo
The early Drosophila embryo is patterned along its anterio-posterior axis by a maternal morphogen, which forms a rouhgly exponential gradient within the embryo. However, due to fluctuations in embryo size, diffusion rates and chemical reaction rates, the gradient of this morphogen (called Bicoid) is not very precise, but shows large variation and it is not sure whether the decay length correlates with embryo size. In contrast, the first downstream target of Bicoid, called hunchback, shows a very precise and scaled gradient, which indicates that more than a simple transcription threshold is at work.
From general considerations, we could show using data on the Bicoid gradient, that a simple model involving a correlated, opposing gradient cannot account for the increased precision observed in the hunchback gradient. On the other hand, we have simulated a reaction diffusion model, which relies on the increased transport of hunchback RNA by a protein present at both ends of the embryo (called staufen). In a very robust manner, this model yields highly precise and scaled hunchback gradients as is observed in the experiment (see figure below). In addition, genetic mutants disrupting the production of staufen are capable of destroying the observation of a precise and scaled hunchback gradient.
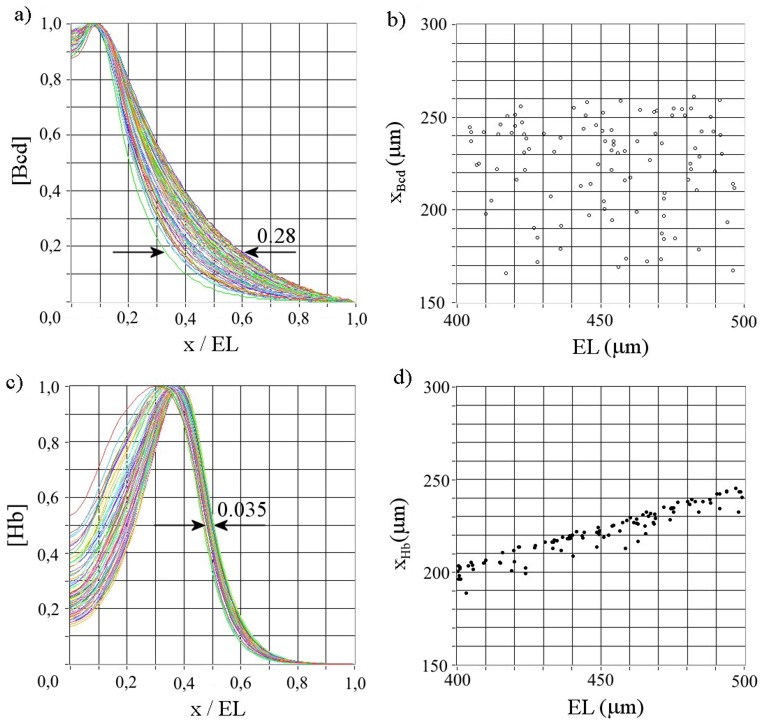
Simulation outcome of a reaction diffusion model of the hunchback gradient formation. While the Bicoid gradient shows large variability in threshold position, the Hunchback gradient is not only precise, but also scales with embryo size.
This project is carried out in collaboration with Tinri Aegerter-Wilmsen. For more information, consult the 2005 J. theor. Biol. paper in the publications section.